Abstract
Objective:
Fanconi anaemia is a genotypically heterogeneous disease. A fast and reliable genetic diagnosis method helps is important for the clinical care of these patients. The objective of this study was to establish a strategy for expeditious molecular diagnosis for the Indian FA patients.
Methods:
Exome sequencing was performed for 119 FA patients on Illumina HiSeq X system and the data was analysed by Sentieon (v201808.01) to identify germline variants. Single nucleotide variants (SNVs) with an allele frequency of <3% in various population databases including the Indian population database were analysed and annotated using an in-house pipeline. Copy number variants (CNVs) were detected using ExomeDepth (v1.1.10). Long amplicon next-generation sequencing was performed for FANCA and FANCG genes. Gene dosage analysis was performed for the deletions identified by WES in the FANCT/UBE2T gene. Sanger sequencing was performed for the selected variants identified by WES.
Results:
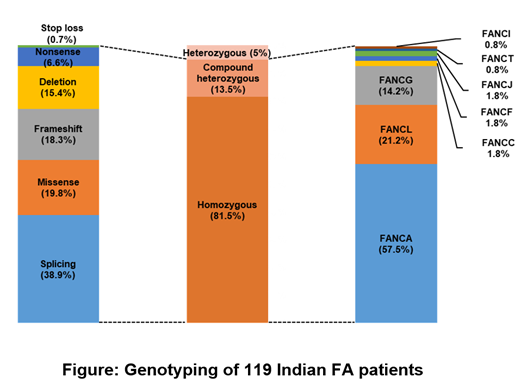
SNVs associated with FA were identified in heterozygous states in 93 (78%) patients. Of the remaining 20 patients, 8 had homozygous deletions, and twelve were compound heterozygous with deletions and SNVs. We detected deletions in 20 patients: 7 with heterozygous and 12 with homozygous deletions in FANCA, and one with a homozygous deletion in FANCT (UBE2T). MLPA confirmed the deletions in the FANCA gene. The homozygous deletion in the FANCT gene was confirmed by the lack of amplification by PCR using the primers binding to the deleted region.
By combining the SNVs and deletions, disease-causing genotypes were identified in 113 of 119 (95%) patients. A large number of our patients (81.5 %) were homozygous (Figure) due to the high rate of consanguinity in the population. FANCA was found to be the most frequently mutated gene in (57.5%) while mutations in FANCG gene accounted for 14.2% of our patients. FANCC mutations were found at a very low frequency (1.8%) in our patients.
Although mutations in FANCL are rare in all the populations, 24 (21.2%) patients with FANCL mutations were identified in our study. A previously reported synonymous splicing mutation
FANCL c.1092G>A;p.K364= was found in homozygous state in 22 (19.5%) patients. Two other FANCL mutations identified includes a missense mutation c.827C>T; p.Pro281Leu in the homozygous state in one patient and a nonsense mutation c.997C>T; p.Gln333Ter in the compound heterozygous state with the common FANCL c.1092G>A;p.K364= mutation in another patient. Mutations in rarely mutated FA genes included 1 patient each in FANCT/UBE2T and FANCI and two each in FANCJ/BRIP1 and FANCF gene. Based on the comprehensive genotype analysis, we could design an algorithm for FA in the Indian population. ~20% of the FA patients who have FANCL c.1092G>A;p.K364= mutation can be diagnosed by Sanger sequencing. MLPA can detect FANCA deletions (16.5% of the overall mutations). The results from these two tests can be obtained in 48 hours. For those who are negative for the mutations by these two methods, LA-NGS can detect SNVs in the FANCA and FANCG genes, which constitute >55% of the FA mutations in the population. We have tested this algorithm for expedited molecular diagnosis in 27 FA patients and found that the disease genotypes could be established in 94% of the patients in less than two weeks.
Conclusion/Clinical applicability:
Exome sequencing identified several novel mutations in the FA pathway genes in the FA patients. A cost-effective and time-saving algorithm was explicitly established for molecular diagnosis of FA in the Indian population.
No relevant conflicts of interest to declare.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal